Parallel and Cluster Computing with R
Elizabeth Byerly
2016-09-27
Press "s" for presenter's notes
Introduction
Making coffee

Single thread

Parallel

Cluster

R is a single-threaded program.
We will learn how to write parallel R programs.
Faster process means more iterations, testing, and experimentation
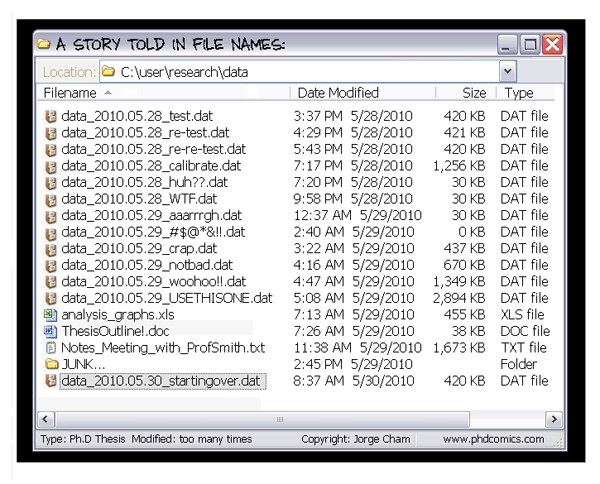
Intractable problems become tractable
Outcomes
Learn jargon and fundamental concepts of parallel programming
Identify parallelizable tasks
Learn base R's parallel syntax
Introduce cluster computing
Agenda
-
Parallel programming
-
Parallel programming in R
-
Cluster computing
Parallel Programming
Parallelizable problems
Work can be split into independent processes:
-
Bootstrapping
-
Random forests
-
Tuning parameters
-
Graphing
-
Data cleaning
-
...
Process independence
-
A process does not rely on another process's outputs
-
Processes do not need to communicate state during execution
Many data, one task.
One data, many tasks.
Parallel overhead
-
Load balancing
-
Communication speed






Parallel Programming in R
The parallel package
-
Base R package
-
Shared syntax with R's functional programming utilities
(the apply functions)
-
Systems for all basic parallel operations
Crash Course: lapply
lapply
returns a list of the same length as X, each element of which is the
result of applying FUN to the corresponding element of X.
- CRAN
example_list <- list(1:10, 11:100, rep(20, 5))
example_vector <- c(1, 3, 16)
lapply(example_list, identity)
## [[1]]
## [1] 1 2 3 4 5 6 7 8 9 10
##
## [[2]]
## [1] 11 12 13 14 15 16 17 18 19 20 21 22 23
## [14] 24 25 26 27 28 29 30 31 32 33 34 35 36
## [27] 37 38 39 40 41 42 43 44 45 46 47 48 49
## [40] 50 51 52 53 54 55 56 57 58 59 60 61 62
## [53] 63 64 65 66 67 68 69 70 71 72 73 74 75
## [66] 76 77 78 79 80 81 82 83 84 85 86 87 88
## [79] 89 90 91 92 93 94 95 96 97 98 99 100
##
## [[3]]
## [1] 20 20 20 20 20
lapply(example_list, mean)
## [[1]]
## [1] 5.5
##
## [[2]]
## [1] 55.5
##
## [[3]]
## [1] 20
lapply(example_vector, identity)
## [[1]]
## [1] 1
##
## [[2]]
## [1] 3
##
## [[3]]
## [1] 16
lapply(example_vector, function(x) x * x)
## [[1]]
## [1] 1
##
## [[2]]
## [1] 9
##
## [[3]]
## [1] 256
lapply(c(identity, mean, sum), function(current_func) {
current_func(example_vector)
})
## [[1]]
## [1] 1 3 16
##
## [[2]]
## [1] 6.666667
##
## [[3]]
## [1] 20
Our First Parallel Program
Bootstrapping an estimate
-
Define a bootstrap for the iris dataset
-
Run the bootstrap function in a single thread
-
Configure a minimal R parallel computing environment
-
Run the bootstrap function in parallel
Our bootstrap function
run_iris_boot <- function(...) {
iris_boot_sample <- iris[sample(1:nrow(iris), replace = TRUE),]
lm(Sepal.Length ~ Sepal.Width + Petal.Length,
data = iris_boot_sample)
}
Run once
run_iris_boot()
## Call:
## lm(formula = Sepal.Length ~ Sepal.Width + Petal.Length,
## data = iris_boot_sample)
##
## Coefficients:
## (Intercept) Sepal.Width Petal.Length
## 2.0756 0.6162 0.4983
Single threaded
set.seed(20160927)
system.time(lapply(1:1000, run_iris_boot))
## user system elapsed
## 1.01 0.00 1.02
Parallel
library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
system.time(parLapply(cluster, 1:1000, run_iris_boot))
## user system elapsed
## 0.19 0.03 0.77
stopCluster(cluster)
Improvement
0.77 / 1.02
## [1] 0.754902
Additional overhead
system.time({
library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)
})
## user system elapsed
## 0.17 0.05 1.35
1.35 / 1.02
## [1] 1.323529
Breaking Down the Example
library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)

library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)

library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)
library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)

library(parallel)
cores <- detectCores()
cluster <- makeCluster(cores)
clusterSetRNGStream(cluster, 20160927)
parLapply(cluster, 1:1000, run_iris_boot)
stopCluster(cluster)

Doing Something Useful
Tuning Parameters
Testing k-means groups
-
Generate and configure our cluster
-
Instruct our worker nodes to load a needed package
-
Run k-means against different potential group counts on our worker
nodes
-
Return the results to our manager session
-
Summarize the fit for different groups in our manager session
clusterEvalQ(cluster, library(MASS))
test_centers <- 2:6
node_results <- parSapply(cluster, test_centers, function(n_centers) {
kmeans(anorexia[2:3], centers = n_centers, nstart = 200)$betweenss
})
final_results <- by(node_results, test_centers, median)
clusterEvalQ(cluster, library(MASS))
test_centers <- 2:6
node_results <- parSapply(cluster, test_centers, function(n_centers) {
kmeans(anorexia[2:3], centers = n_centers, nstart = 200)$betweenss
})
final_results <- by(node_results, test_centers, median)
clusterEvalQ(cluster, library(MASS))
test_centers <- 2:6
node_results <- parSapply(cluster, test_centers, function(n_centers) {
kmeans(anorexia[2:3], centers = n_centers, nstart = 200)$betweenss
})
final_results <- by(node_results, test_centers, median)
clusterEvalQ(cluster, library(MASS))
test_centers <- 2:6
node_results <- parSapply(cluster, test_centers, function(n_centers) {
kmeans(anorexia[2:3], centers = n_centers, nstart = 200)$betweenss
})
final_results <- by(node_results, test_centers, median)
clusterEvalQ(cluster, library(MASS))
test_centers <- 2:6
node_results <- parSapply(cluster, test_centers, function(n_centers) {
kmeans(anorexia[2:3], centers = n_centers, nstart = 200)$betweenss
})
final_results <- by(node_results, test_centers, median)
Doing Something Useful
Cleaning Data
Cleaning text data
-
Generate and configure our cluster
-
Export database information to our worker nodes
-
Instruct our worker nodes to create a connection to the database
-
Split the raw text file paths across our worker nodes and instruct
them to read each text file, clean the data, and write it to the
database
clusterExport(cluster, c("db_user", "db_password", "db_host"))
clusterEvalQ(cluster, {
db_conn <- dbConnect(user=db_user, password=db_password,
host=db_host)
})
parLapply(cluster, raw_data_files, function(file_path) {
df <- read.csv(file_path)
df$y <- toupper(df$y)
dbWriteTable(db_conn, df, )
})
db_user <- "elizabeth"
db_user
## [1] "elizabeth"
clusterEvalQ(cluster, db_user)
## Error in checkForRemoteErrors(lapply(cl, recvResult)) :
## 2 nodes produced errors; first error: object 'db_user' not found
clusterExport(cluster, c("db_user"))
clusterEvalQ(cluster, db_user)
## [[1]]
## [1] "elizabeth"
##
## [[2]]
## [1] "elizabeth"
clusterExport(cluster, c("db_user", "db_password", "db_host"))
clusterEvalQ(cluster, {
db_conn <- dbConnect(user=db_user, password=db_password,
host=db_host)
})
parLapply(cluster, raw_data_files, function(file_path) {
df <- read.csv(file_path)
df$y <- toupper(df$y)
dbWriteTable(db_conn, df, )
})
clusterExport(cluster, c("db_user", "db_password", "db_host"))
clusterEvalQ(cluster, {
db_conn <- dbConnect(user=db_user, password=db_password,
host=db_host)
})
parLapply(cluster, raw_data_files, function(file_path) {
df <- read.csv(file_path)
df$y <- toupper(df$y)
dbWriteTable(db_conn, df, )
})
clusterExport(cluster, c("db_user", "db_password", "db_host"))
clusterEvalQ(cluster, {
db_conn <- dbConnect(user=db_user, password=db_password,
host=db_host)
})
parLapply(cluster, raw_data_files, function(file_path) {
df <- read.csv(file_path)
df$y <- toupper(df$y)
dbWriteTable(db_conn, df, )
})
clusterExport(cluster, c("db_user", "db_password", "db_host"))
clusterEvalQ(cluster, {
db_conn <- dbConnect(user=db_user, password=db_password,
host=db_host)
})
parLapply(cluster, raw_data_files, function(file_path) {
df <- read.csv(file_path)
df$y <- toupper(df$y)
dbWriteTable(db_conn, df, )
})
Evaluating performance
When does it make sense to absorb the parallel overhead?
-
Many computations against the same data
-
The same computation against many data
-
No need to communicate mid-process
Check your assumptions using system.time()
Monitoring nodes
Error messages are typically obtuse.
## Error in checkForRemoteErrors(val) :
## 4 nodes produced errors; first error: 1
-
Test your code in a single-threaded session
-
Create log files for worker nodes
Random number generators
clusterSetRNGStream(cluster, 20160927)
Two things to consider:
-
For L'Ecuyer to work, the same number of worker processes must be
fed streams
-
We cannot reproduce the results in a single-threaded process
A cluster is a number of computers configured to work together across a
network as a single system for some task.

For our purposes, a cluster is computers running local R sessions that
take commands and return outputs to a manager session.

makePSOCKCluster()
-
Given a number, make that many local Rscript sessions.
-
Given a character vector, use each value as a network address and
instantiate a remote Rscript session.
-
The manager R session tracks the network location and port of each
Rscript worker node.
-
Rscript worker nodes listen on a port for instructions and serial
data from the manager.
Computers and networks default to closed traffic for security.

Minimum networking knowledge
-
SSH, secure communication across networks
-
Firewalls, traffic control at network boundaries
-
Ports, computers listening for traffic from the network
Steps
-
Launch two computers on a secure network
-
Install the necessary software on both computers
-
Share the private network IPs and SSH credentials across the two
computers
-
Use makePSOCKcluster() and the private
network IP addresses to create worker node R sessions
Launch two computers

Install the necessary software

sudo apt-get install r-base-dev openssh-server
Share IPs and SSH credentials

Create a worker node R session

Create a worker node R session
library(parallel)
cluster <- makePSOCKcluster(c("172.31.51.171", "localhost"))
clusterEvalQ(cluster, {
system("ifconfig eth0 | grep 'inet addr' | awk '{print $2}'")
})
## addr:172.31.50.241
## addr:172.31.51.171